Single Cell Technologies and Analysis: Seurat and friends
2025-02-14
Why Single Cell?
 Identify biomarkers of cell types and states
Identify biomarkers of cell types and states
 Capture full heterogeneity in a sample
Capture full heterogeneity in a sample
 Track changes in states: development, disease progression, immune responses
Track changes in states: development, disease progression, immune responses
 Explore cell-to-cell interactions
Explore cell-to-cell interactions
How to choose a method?
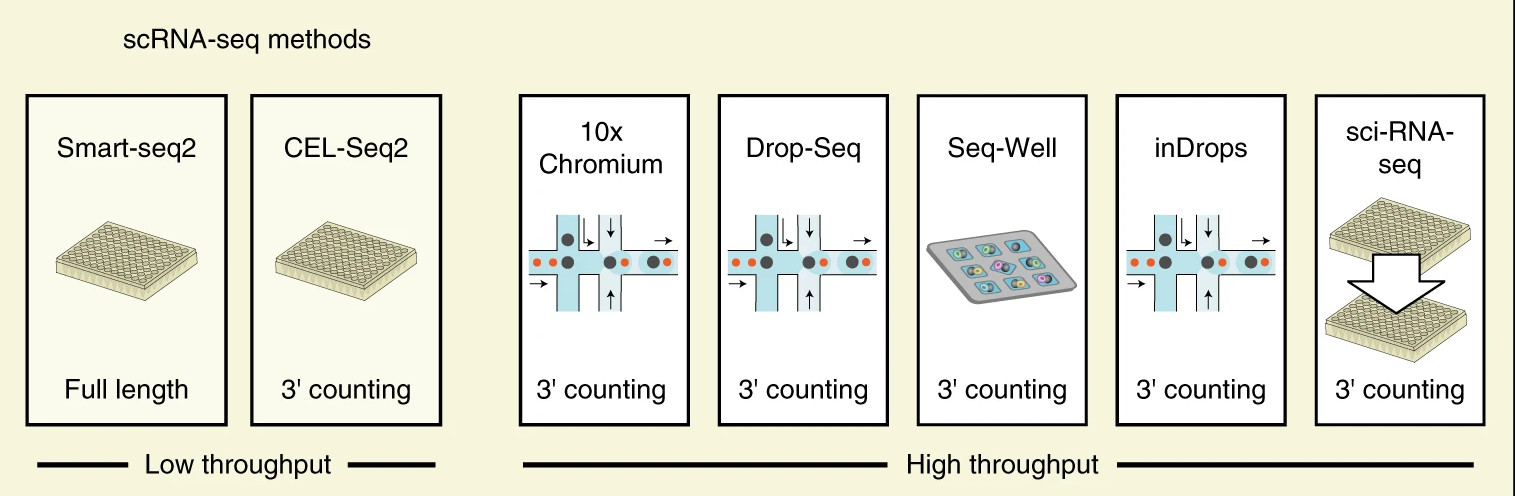
Ding, J. et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat Biotechnol 38, 737–746 (2020).
How to choose a method? Number of UMI / Genes
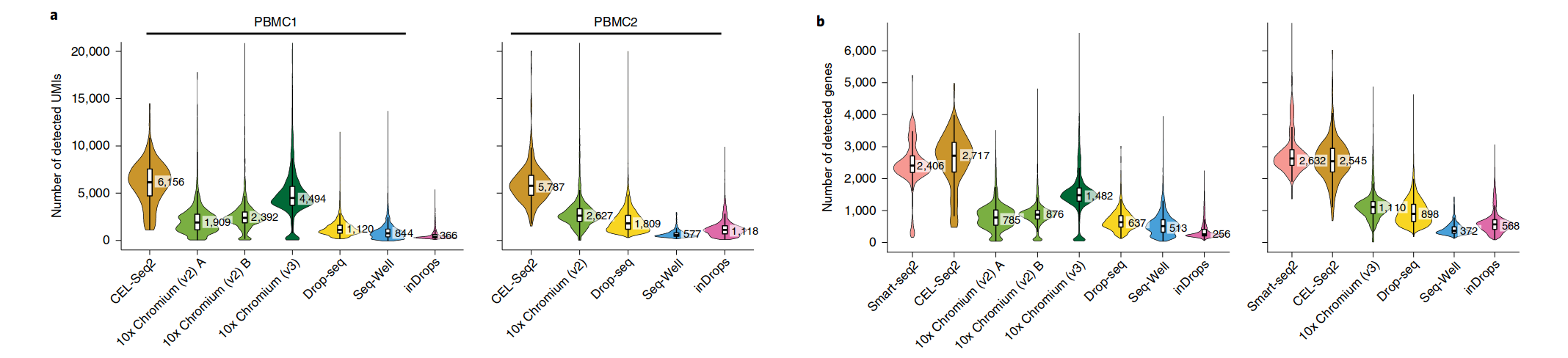
Ding, J. et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat Biotechnol 38, 737–746 (2020).
How to choose a method: considerations
10x Chromium had the strongest consistent performance: high sensitivity, fastest
Lowest costs: Drop-seq, Seq-Well and inDrops
High sensitivity: both low-throughput methods (Smart-seq2 and CEL-Seq2)
Smart-seq2: full-length transcript allows for splicing isoforms and variant detection
Ding, J. et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat Biotechnol 38, 737–746 (2020).
scRNA-seq computational pre-processing
For 10X: Cell Ranger (https://www.10xgenomics.com/support/software/cell-ranger/latest/algorithms-overview/cr-3p-cellplex-algorithm)
Others: Alevin (Salmon), Kallisto BUStools (kb), STARsolo
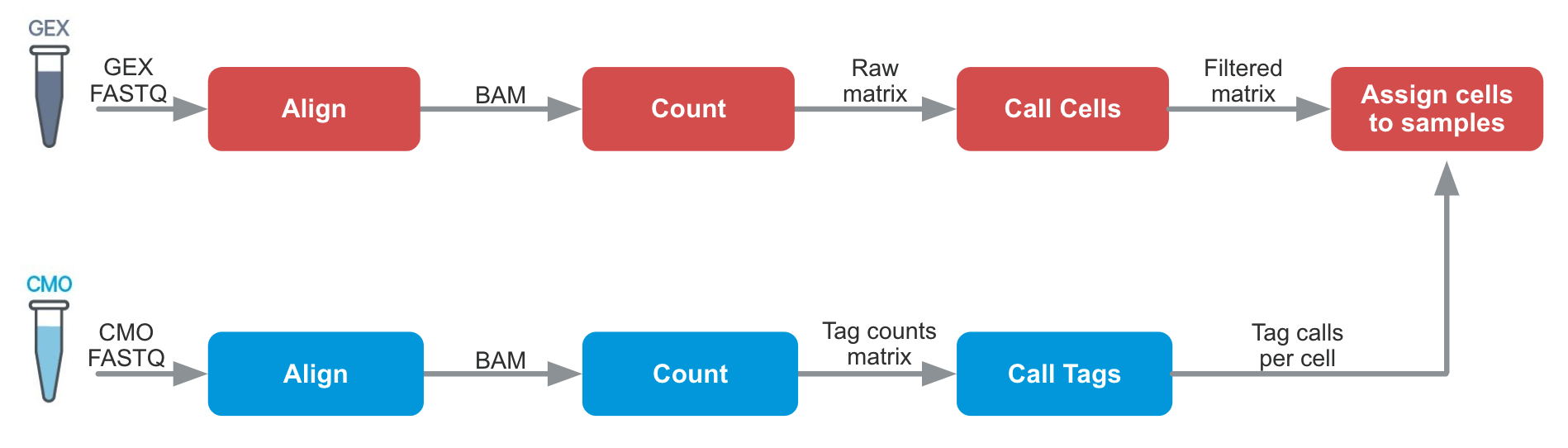
CMO: Cell Multiplexing Oligo tags
GEX: Gene expression
cellranger demultiplexing
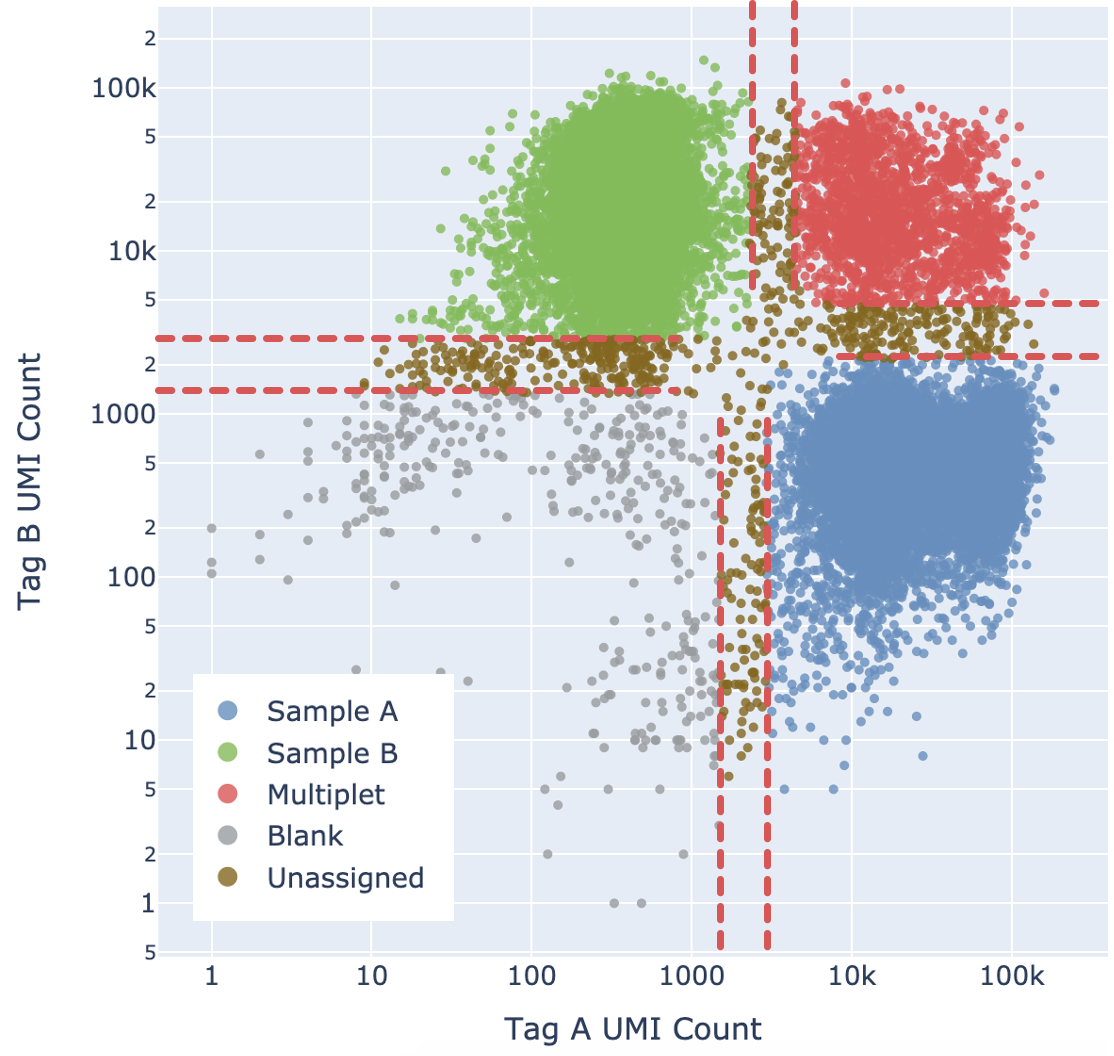
Expectation-Maximization (EM) algorithm of the parameters of the Gaussians and state assignments
New! cellranger annotate
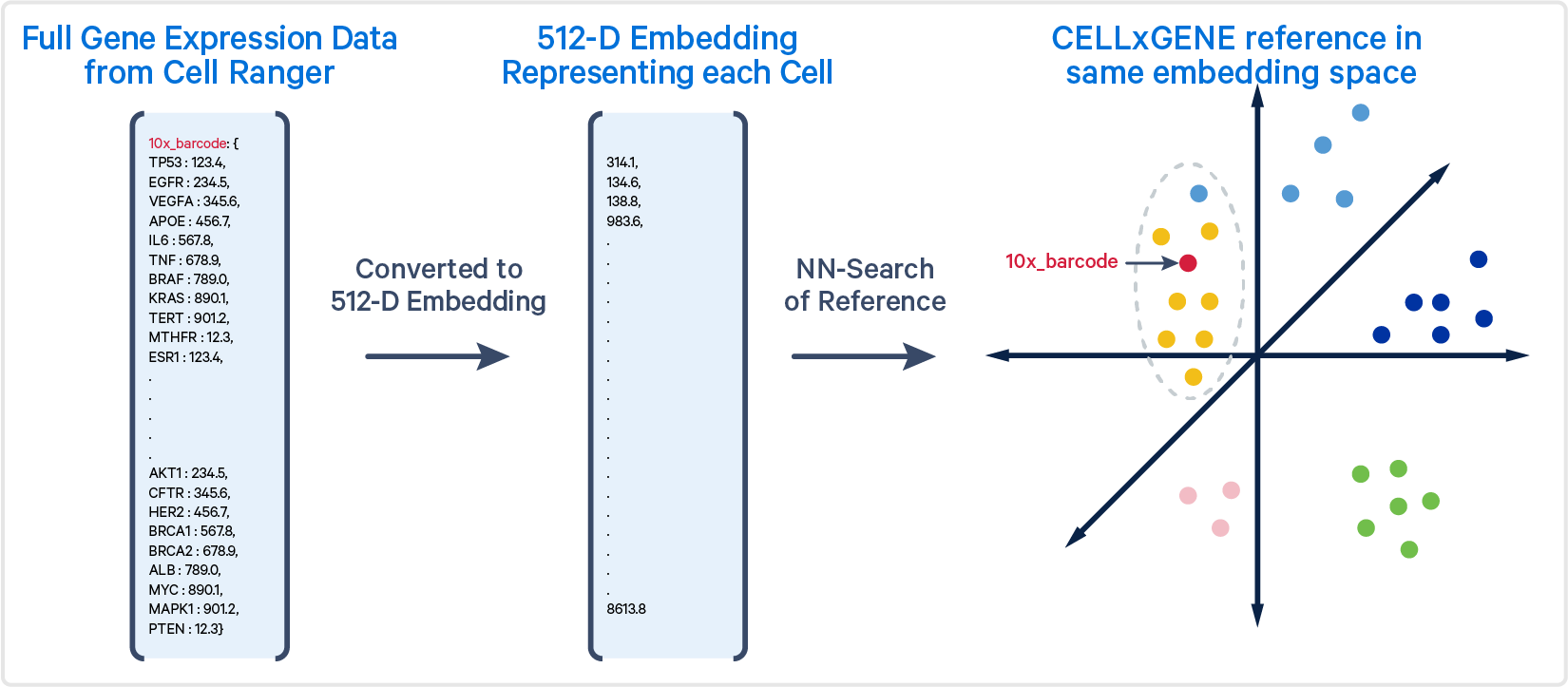
Analysis library families



| Seurat | Bioconductor | scverse | |
|---|---|---|---|
| Programming Language | R | R | Python |
| Core Data Structure | Seurat object | SingleCellExperiment | AnnData |
| Ecosystem | Integrated all-in-one “pipeline” | Modular via Bioconductor packages | Modular, interoperable toolkit |
| Visualization | Extensive built-in plotting functions | Visualization via scatter + ggplot2 | scanpy + MPL / seaborn |
| Community & Support | Large, active community | Bioconductor project | Growing, active Python 3community |
State-of-the art single-cell with Seurat
Follow me to the tutorial: csgroen.github.io/posts/tutorial_seurat_du/